pacman::p_load(seriation, dendextend, heatmaply, tidyverse)Hands-on Exercise 5c: Heatmap for Visualising and Analysing Multivariate Data
1 Getting Started
In this exercise, we will use seriation, heatmaply, dendextend and tidyverse in RStudio.
The code chunk below uses p_load() of pacman package to check if these packages are installed in the computer and load them onto your working R environment.
The code chunk below imports the data into R environment by using read_csv() function of readr package.
wh <- read_csv("data/WHData-2018.csv")We need to change the rows by country name instead of row number by using the code chunk below
row.names(wh) <- wh$CountryThe code chunk below will be used to transform wh data frame into a data matrix.
wh1 <- dplyr::select(wh, c(3, 7:12))
wh_matrix <- data.matrix(wh)2 Static Heatmap
There are many R packages and functions can be used to drawing static heatmaps, they are:
heatmap()of R stats package. It draws a simple heatmap.
heatmap.2() of gplots R package. It draws an enhanced heatmap compared to the R base function.
pheatmap() of pheatmap R package. pheatmap package also known as Pretty Heatmap. The package provides functions to draws pretty heatmaps and provides more control to change the appearance of heatmaps.
ComplexHeatmap package of R/Bioconductor package. The package draws, annotates and arranges complex heatmaps (very useful for genomic data analysis). The full reference guide of the package is available here.
superheat package: A Graphical Tool for Exploring Complex Datasets Using Heatmaps. A system for generating extendable and customizable heatmaps for exploring complex datasets, including big data and data with multiple data types. The full reference guide of the package is available here.
In this section, we will use heatmap() of R Stats package to plot static heatmaps.
2.1 heatmap() of R Stats
We will plot a heatmap by using heatmap() of Base Stats. The code chunk is given below.
wh_heatmap <- heatmap(wh_matrix,
Rowv=NA, Colv=NA)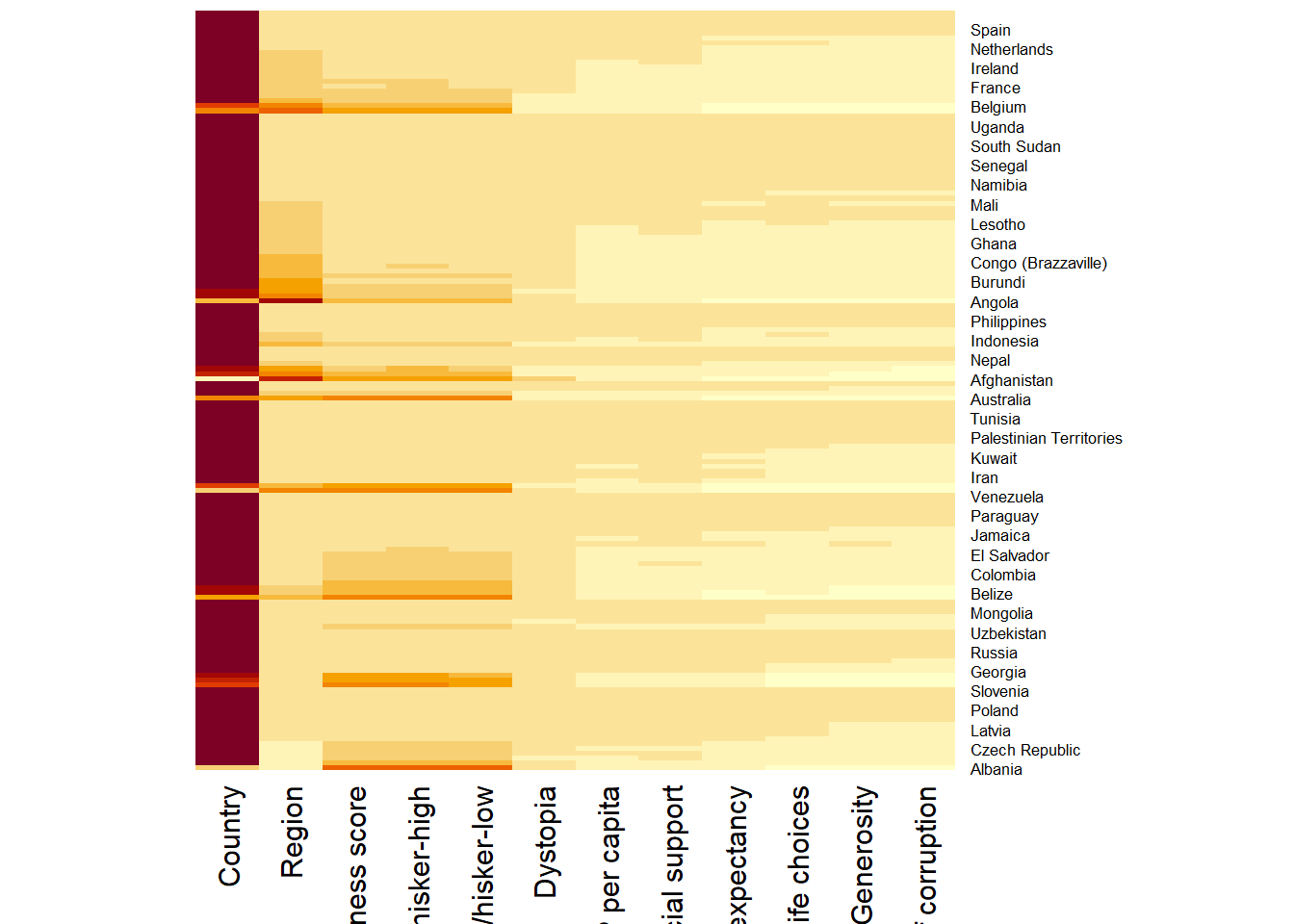
- By default, heatmap() plots a cluster heatmap. The arguments Rowv=NA and Colv=NA are used to switch off the option of plotting the row and column dendrograms.
To plot a cluster heatmap, we just have to use the default as shown in the code chunk below.
wh_heatmap <- heatmap(wh_matrix)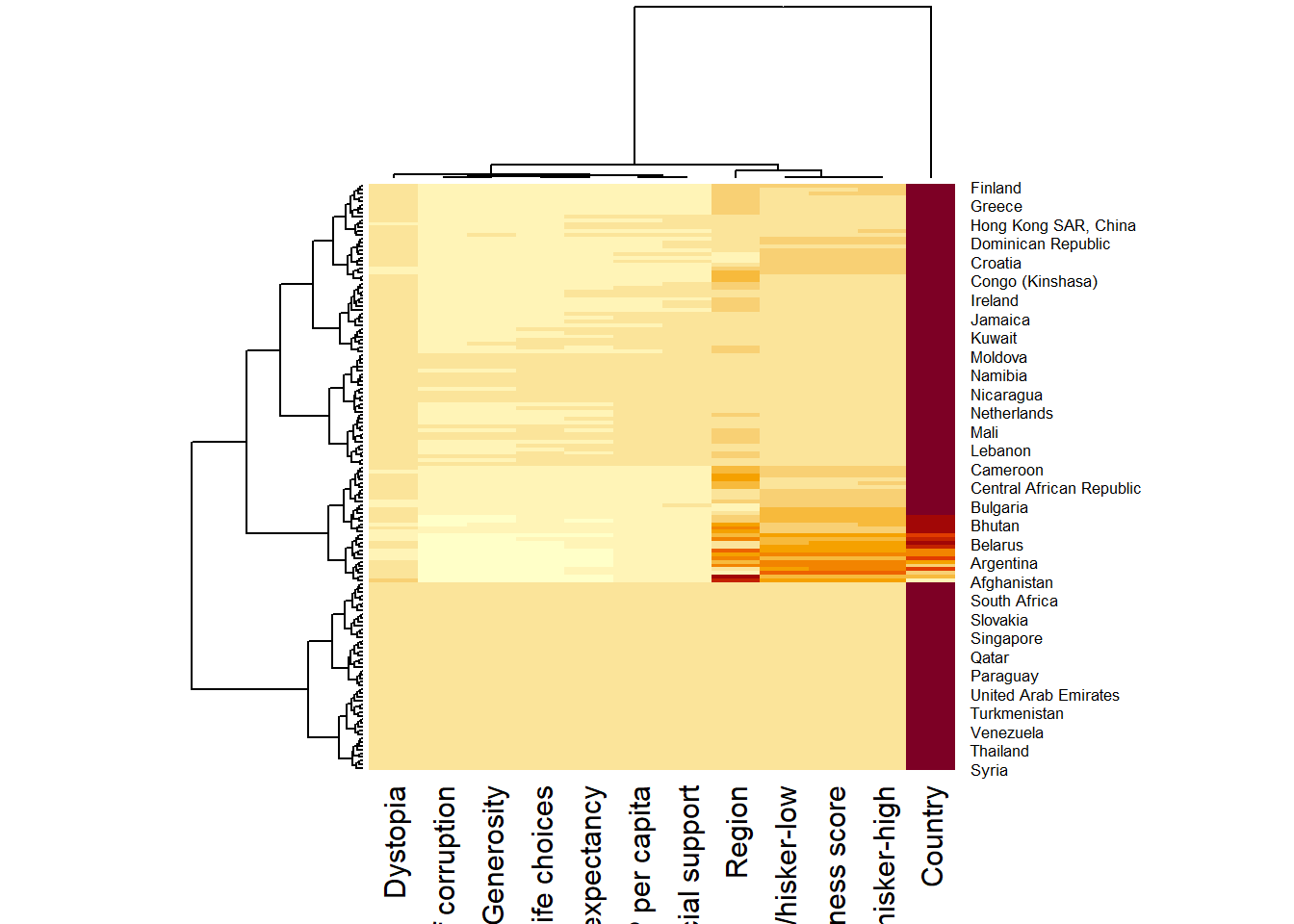
- The order of both rows and columns is different compare to the native wh_matrix. This is because heatmap do a reordering using clusterisation: it calculates the distance between each pair of rows and columns and try to order them by similarity. Moreover, the corresponding dendrogram are provided beside the heatmap.
The code chunk below normalizes the matrix column-wise.
wh_heatmap <- heatmap(wh_matrix,
scale="column",
cexRow = 0.6,
cexCol = 0.8,
margins = c(10, 4))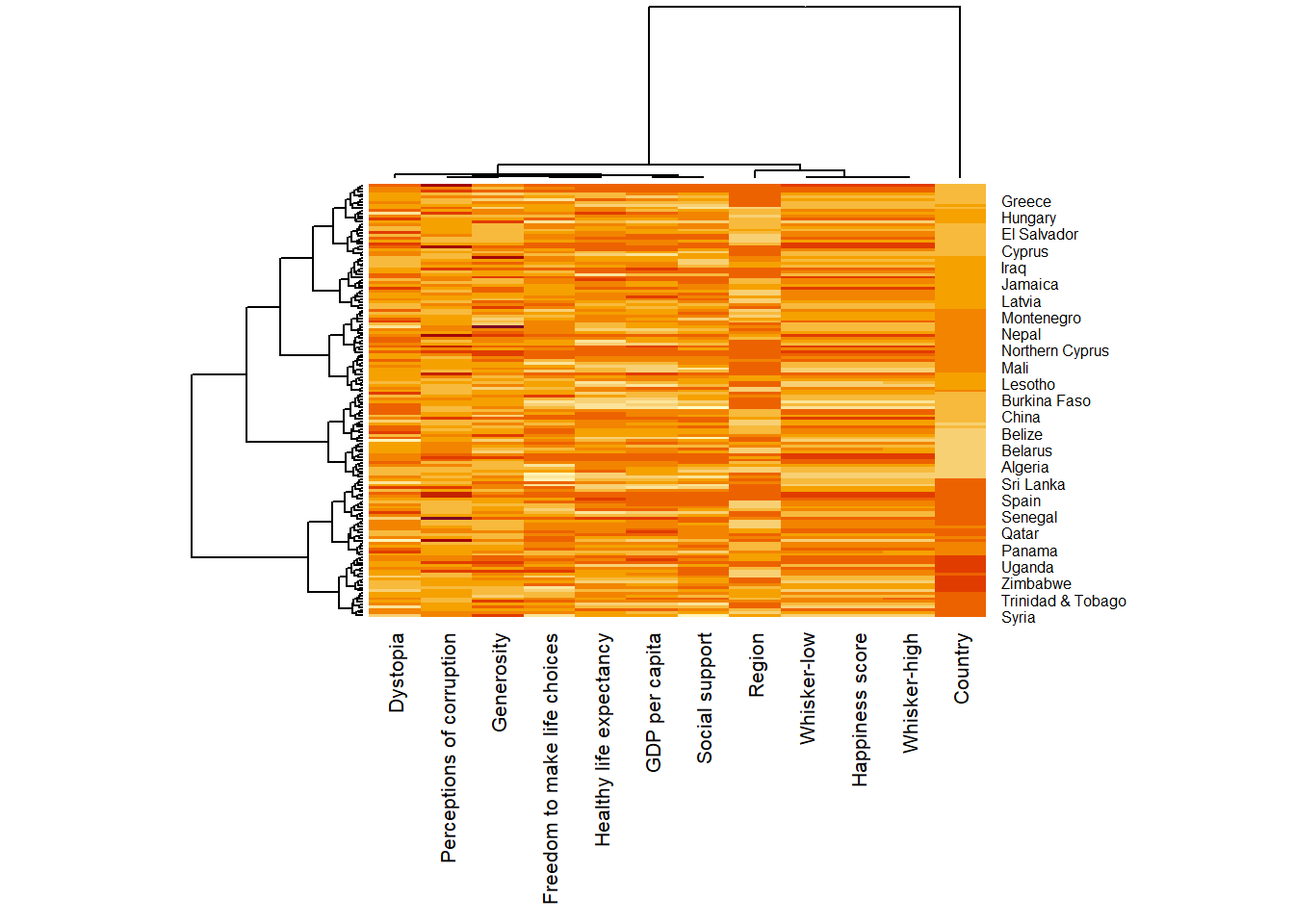
- Notice that the values are scaled now. Also note that margins argument is used to ensure that the entire x-axis labels are displayed completely and, cexRow and cexCol arguments are used to define the font size used for y-axis and x-axis labels respectively.
3 Creating Interactive Heatmap
heatmaply is an R package for building interactive cluster heatmap that can be shared online as a stand-alone HTML file.
3.1 Working with heatmaply
heatmaply(mtcars)The code chunk below shows the basic syntax needed to create n interactive heatmap by using heatmaply package.
heatmaply(wh_matrix[, -c(1, 2, 4, 5)],
cexRow = 0.4,
cexCol = 0.8,
margins = c(10, 10))- Different from heatmap(), for heatmaply() the default horizontal dendrogram is placed on the left hand side of the heatmap.
- The text label of each raw, on the other hand, is placed on the right hand side of the heat map.
- When the x-axis marker labels are too long, they will be rotated by 135 degree from the north.
3.2 Data trasformation
There are three main data transformation methods supported by heatmaply() which are scale, normalize and percentilse.
3.2.1 Scaling method
The code chunk below is used to scale variable values columewise.
heatmaply(wh_matrix[, -c(1, 2, 4, 5)],
scale = "column")3.2.2 Normalizing method
Different from Scaling, the normalize method is performed on the input data set i.e. wh_matrix as shown in the code chunk below.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]))3.2.3 Percentizing method
Similar to Normalize method, the Percentize method is also performed on the input data set i.e. wh_matrix as shown in the code chunk below.
heatmaply(percentize(wh_matrix[, -c(1, 2, 4, 5)]))3.3 Clustering algorithm
heatmaply supports a variety of hierarchical clustering algorithm. The main arguments provided are:
distfun: function used to compute the distance (dissimilarity) between both rows and columns. Defaults to dist. The options “pearson”, “spearman” and “kendall” can be used to use correlation-based clustering, which uses as.dist(1 - cor(t(x))) as the distance metric (using the specified correlation method).
hclustfun: function used to compute the hierarchical clustering when Rowv or Colv are not dendrograms. Defaults to hclust.
dist_method default is NULL, which results in “euclidean” to be used. It can accept alternative character strings indicating the method to be passed to distfun. By default distfun is “dist”” hence this can be one of “euclidean”, “maximum”, “manhattan”, “canberra”, “binary” or “minkowski”.
hclust_method default is NULL, which results in “complete” method to be used. It can accept alternative character strings indicating the method to be passed to hclustfun. By default hclustfun is hclust hence this can be one of “ward.D”, “ward.D2”, “single”, “complete”, “average” (= UPGMA), “mcquitty” (= WPGMA), “median” (= WPGMC) or “centroid” (= UPGMC).
3.4 Manual approach
In the code chunk below, the heatmap is plotted by using hierachical clustering algorithm with “Euclidean distance” and “ward.D” method
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
dist_method = "euclidean",
hclust_method = "ward.D")3.5 Statistical approach
In order to determine the best clustering method and number of cluster the dend_expend() and find_k() functions of dendextend package will be used.
First, the dend_expend() will be used to determine the recommended clustering method to be used.
wh_d <- dist(normalize(wh_matrix[, -c(1, 2, 4, 5)]), method = "euclidean")
dend_expend(wh_d)[[3]] dist_methods hclust_methods optim
1 unknown ward.D 0.6137851
2 unknown ward.D2 0.6289186
3 unknown single 0.4774362
4 unknown complete 0.6434009
5 unknown average 0.6701688
6 unknown mcquitty 0.5020102
7 unknown median 0.5901833
8 unknown centroid 0.6338734The output table shows that “average” method should be used because it gave the high optimum value.
Next, find_k() is used to determine the optimal number of cluster.
wh_clust <- hclust(wh_d, method = "average")
num_k <- find_k(wh_clust)
plot(num_k)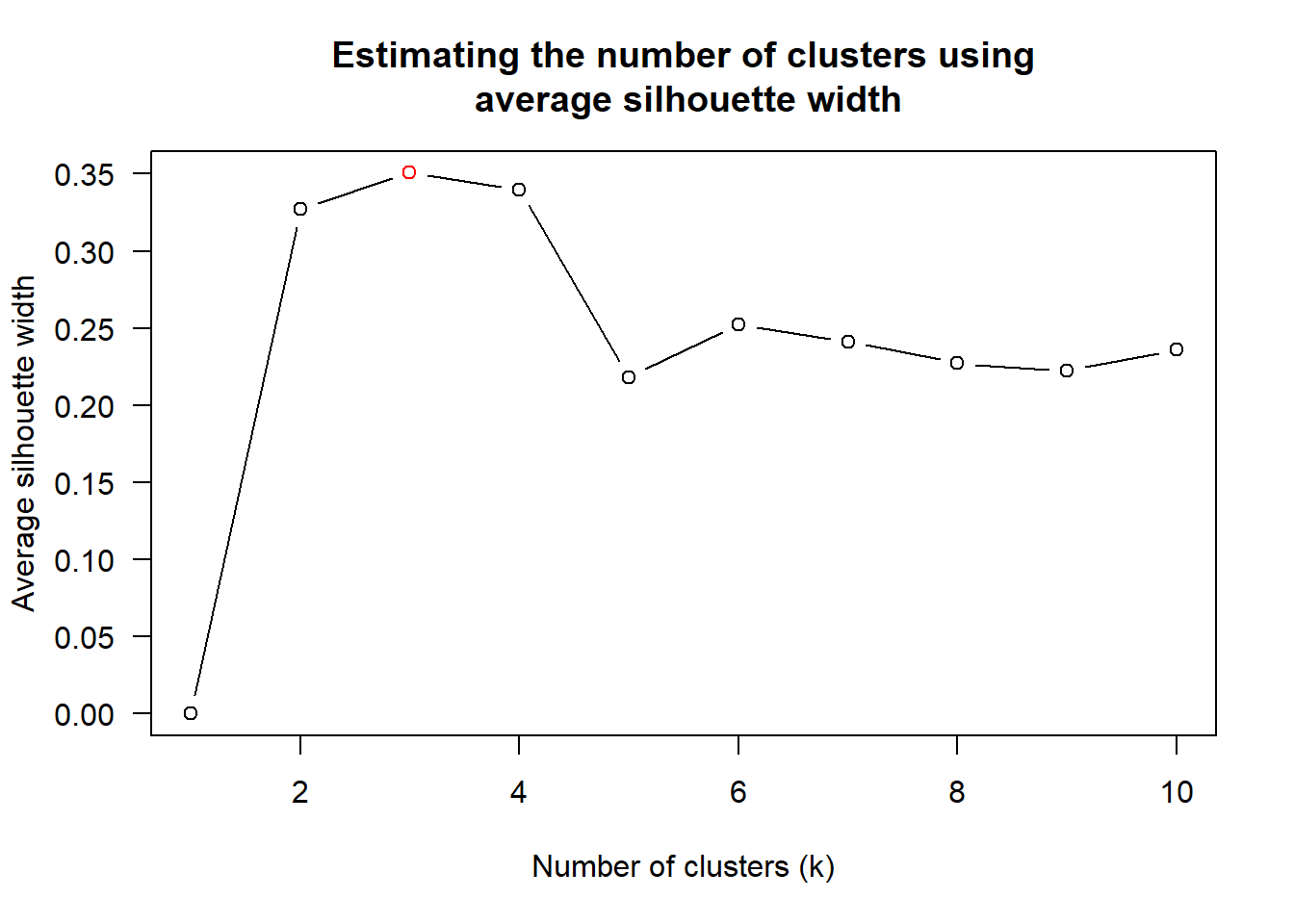
With reference to the statistical analysis results, we can prepare the code chunk as shown below
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
dist_method = "euclidean",
hclust_method = "average",
k_row = 3)3.6 Seriation
heatmaply uses the seriation package to find an optimal ordering of rows and columns. Optimal means to optimize the Hamiltonian path length that is restricted by the dendrogram structure.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "OLO")The default options is “OLO” (Optimal leaf ordering) which optimizes the above criterion (in O(n^4)). Another option is “GW” (Gruvaeus and Wainer) which aims for the same goal but uses a potentially faster heuristic.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "GW")The option “mean” gives the output we would get by default from heatmap functions in other packages such as gplots::heatmap.2.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "mean")The option “none” gives us the dendrograms without any rotation that is based on the data matrix.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "none")3.7 Working with colour palettes
The default colour palette uses by heatmaply is viridis. heatmaply users, however, can use other colour palettes in order to improve the aestheticness and visual friendliness of the heatmap.
In the code chunk below, the Reds color palette of rColorBrewer is used.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "none",
colors = Reds)3.8 The finishing touch
In the code chunk below the following arguments are used:
k_row is used to produce 5 groups.
margins is used to change the top margin to 60 and row margin to 200.
fontsizw_row and fontsize_col are used to change the font size for row and column labels to 4.
main is used to write the main title of the plot.
xlab and ylab are used to write the x-axis and y-axis labels respectively.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
Colv=NA,
seriate = "none",
colors = Reds,
k_row = 5,
margins = c(NA,200,60,NA),
fontsize_row = 4,
fontsize_col = 5,
main="World Happiness Score and Variables by Country, 2018 \nDataTransformation using Normalise Method",
xlab = "World Happiness Indicators",
ylab = "World Countries"
)