pacman::p_load(igraph, tidygraph, ggraph,
visNetwork, lubridate, clock,
tidyverse, graphlayouts)In-class Exercise 9
Loading R package
In this exercise, four network data modelling and visualisation packages will be installed and launched. They are igraph, tidygraph, ggraph and visNetwork. Beside these four packages, tidyverse and lubridate, an R package specially designed to handle and wrangling time data will be installed and launched too.
Importing Network Data from Files
The code chunk below imports GAStech_email_node.csv and GAStech_email_edges-v2.csv into R environment by using read_csv() function of readr package.
GAStech_nodes <- read_csv("data/GAStech_email_node.csv")
GAStech_edges <- read_csv("data/GAStech_email_edge-v2.csv")Data Wrangling
GAStech_edges <- GAStech_edges %>%
mutate(SendDate = dmy(SentDate)) %>%
mutate(Weekday = wday(SentDate,
label = TRUE,
abbr = FALSE))Wrangling Attributes
A close examination of GAStech_edges data.frame reveals that it consists of individual e-mail flow records. This is not very useful for visualisation.
In view of this, we will aggregate the individual by date, senders, receivers, main subject and day of the week.
The code chunk:
GAStech_edges_aggregated <- GAStech_edges %>%
filter(MainSubject == "Work related") %>%
group_by(source, target, Weekday) %>%
summarise(Weight = n()) %>%
filter(source!=target) %>% #avoid those who sending to themselves
filter(Weight > 1) %>%
ungroup()Using tbl_graph() to Build tidygraph Data Model
Use tbl_graph() of tinygraph package to build an tidygraph’s network graph data.frame.
GAStech_graph <- tbl_graph(nodes = GAStech_nodes,
edges = GAStech_edges_aggregated,
directed = TRUE)Changing the Active Object
The nodes tibble data frame is activated by default, but you can change which tibble data frame is active with the activate() function. Thus, if we wanted to rearrange the rows in the edges tibble to list those with the highest “weight” first, we could use activate() and then arrange().
GAStech_graph %>%
activate(edges) %>%
arrange(desc(Weight))# A tbl_graph: 54 nodes and 1372 edges
#
# A directed multigraph with 1 component
#
# Edge Data: 1,372 × 4 (active)
from to Weekday Weight
<int> <int> <ord> <int>
1 40 41 Saturday 13
2 41 43 Monday 11
3 35 31 Tuesday 10
4 40 41 Monday 10
5 40 43 Monday 10
6 36 32 Sunday 9
7 40 43 Saturday 9
8 41 40 Monday 9
9 19 15 Wednesday 8
10 35 38 Tuesday 8
# ℹ 1,362 more rows
#
# Node Data: 54 × 4
id label Department Title
<dbl> <chr> <chr> <chr>
1 1 Mat.Bramar Administration Assistant to CEO
2 2 Anda.Ribera Administration Assistant to CFO
3 3 Rachel.Pantanal Administration Assistant to CIO
# ℹ 51 more rowsPlotting a Basic Network Graph
ggraph(GAStech_graph) +
geom_edge_link() +
geom_node_point() #geom under ggraph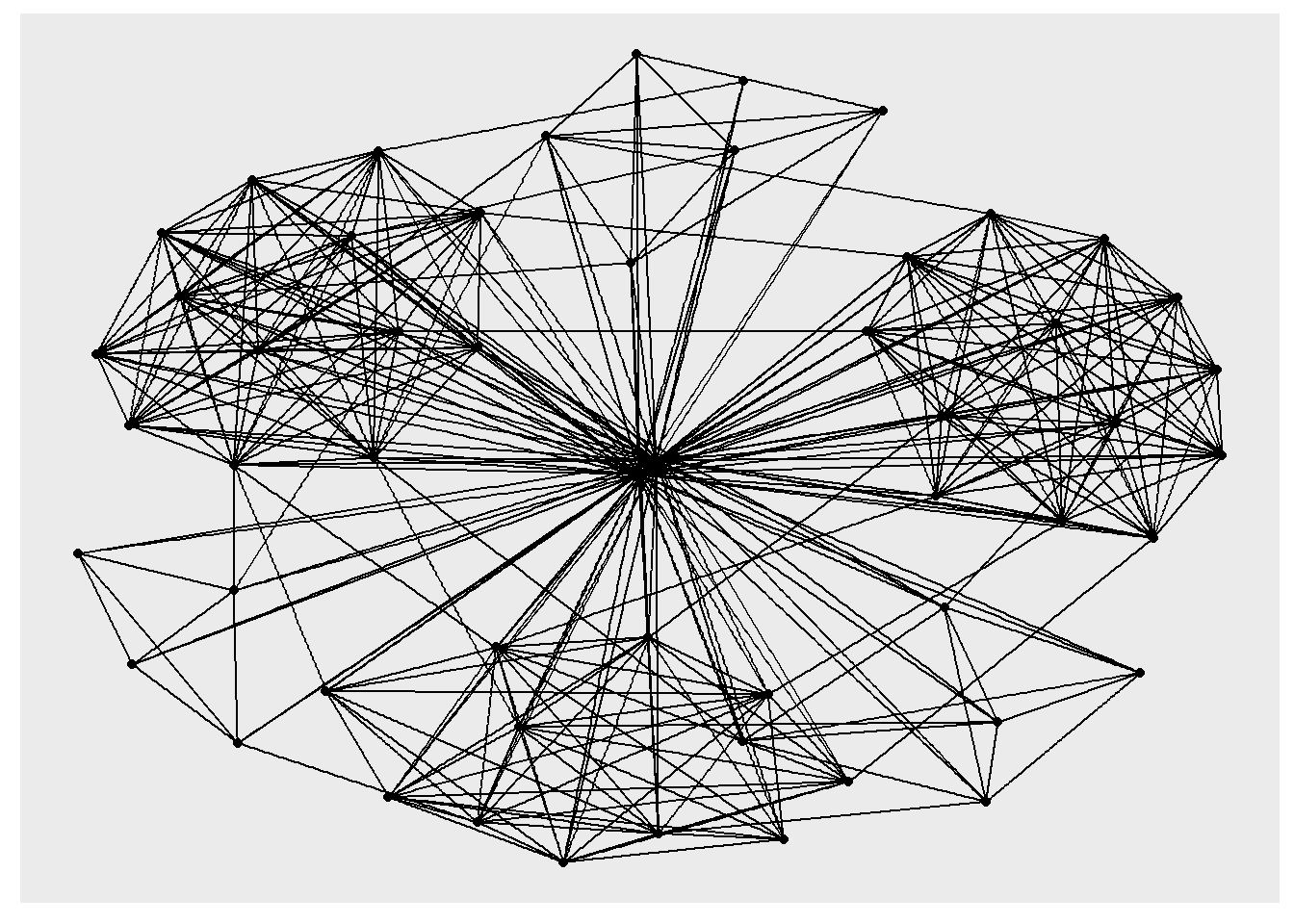
Changing the Default Network Graph Theme
g <- ggraph(GAStech_graph) +
geom_edge_link(aes()) +
geom_node_point(aes())
g + theme_graph()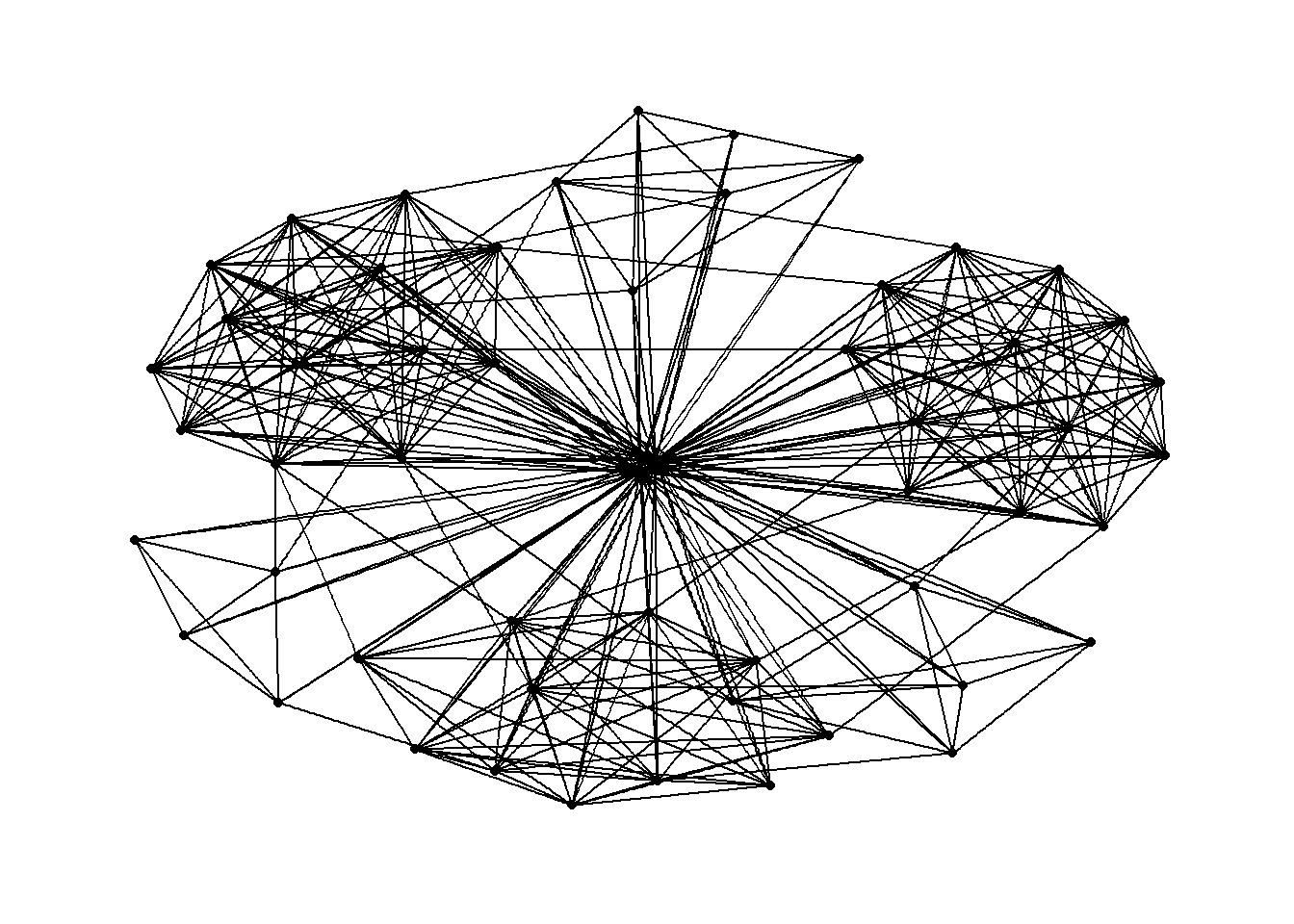
Fruchterman and Reingold Layout
The code chunks below will be used to plot the network graph using Fruchterman and Reingold layout.
g <- ggraph(GAStech_graph,
layout = "fr") +
geom_edge_link(aes()) +
geom_node_point(aes())
g + theme_graph()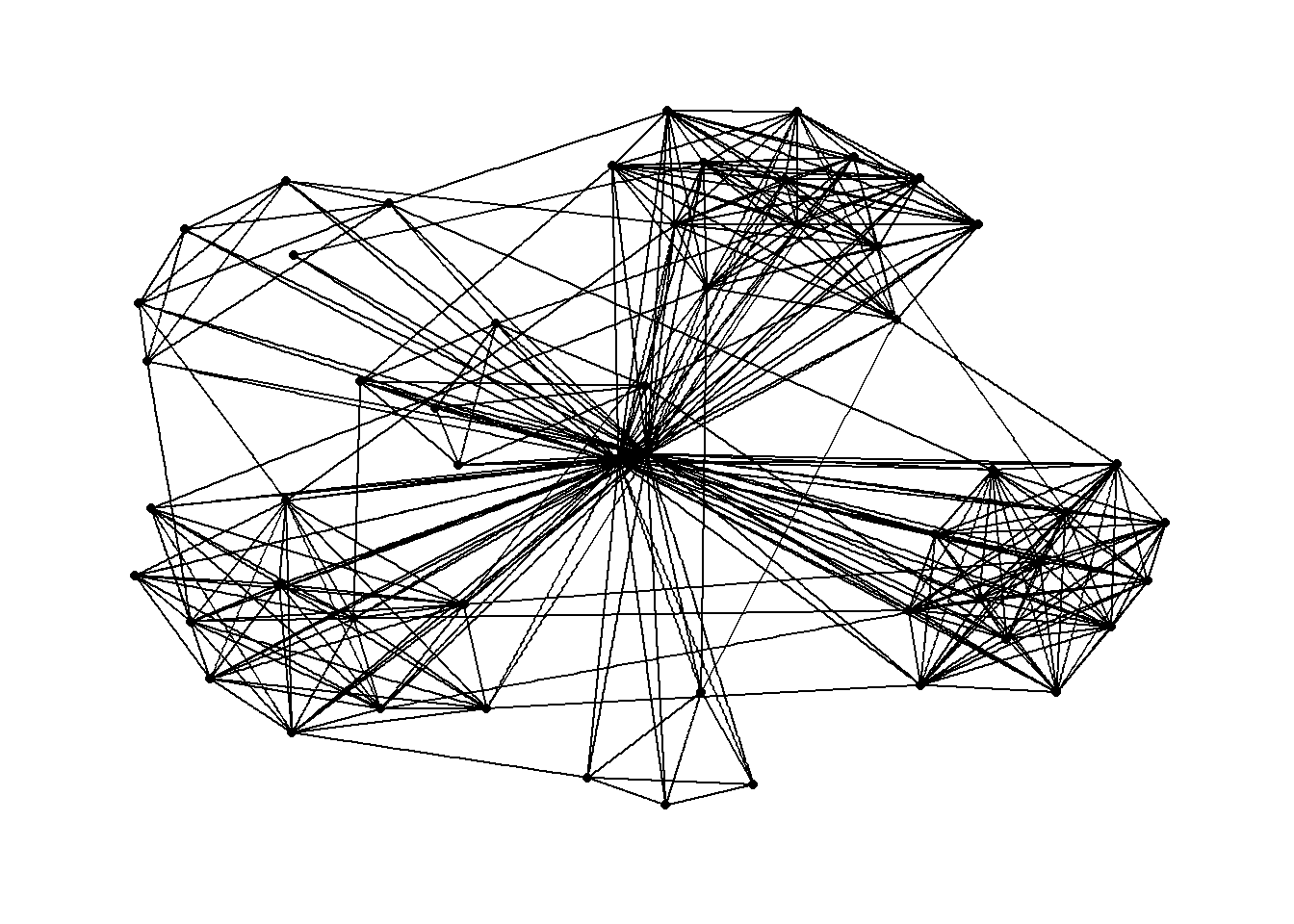
Modifying Network Nodes
g <- ggraph(GAStech_graph,
layout="nicely") +
geom_edge_link(aes()) +
geom_node_point(aes(colour= Department,
size=3))
g + theme_graph()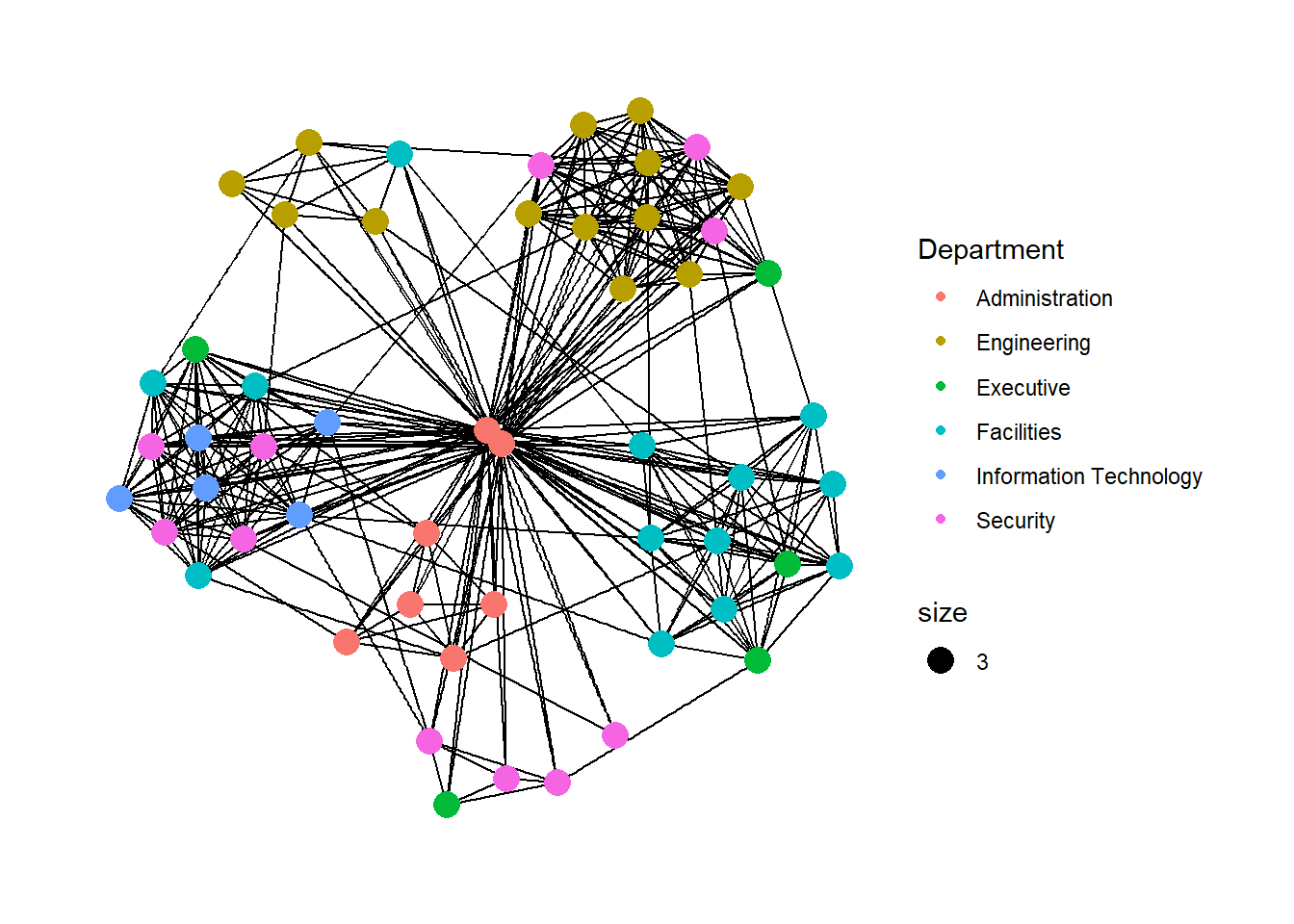
Modifying Edges
g <- ggraph(GAStech_graph,
layout = "nicely") +
geom_edge_link(aes(width=Weight),
alpha=0.2) +
scale_edge_width(range = c(0.1, 5)) +
geom_node_point(aes(colour = Department),
size = 3)
g + theme_graph()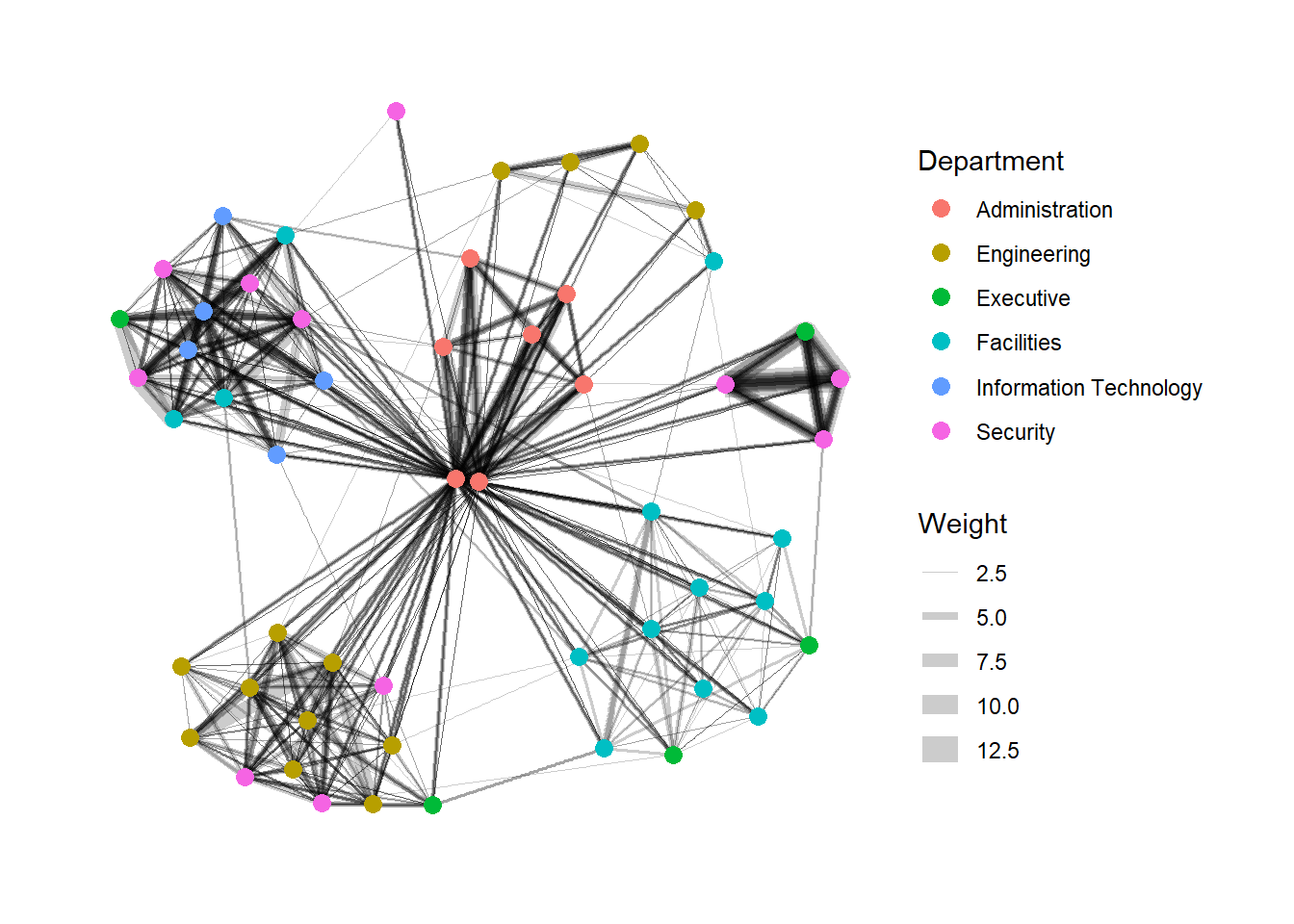
Working with facet_edges()
set_graph_style()
g <- ggraph(GAStech_graph,
layout = "nicely") +
geom_edge_link(aes(width=Weight),
alpha=0.2) +
scale_edge_width(range = c(0.1, 5)) +
geom_node_point(aes(colour = Department),
size = 2)
g + facet_edges(~Weekday)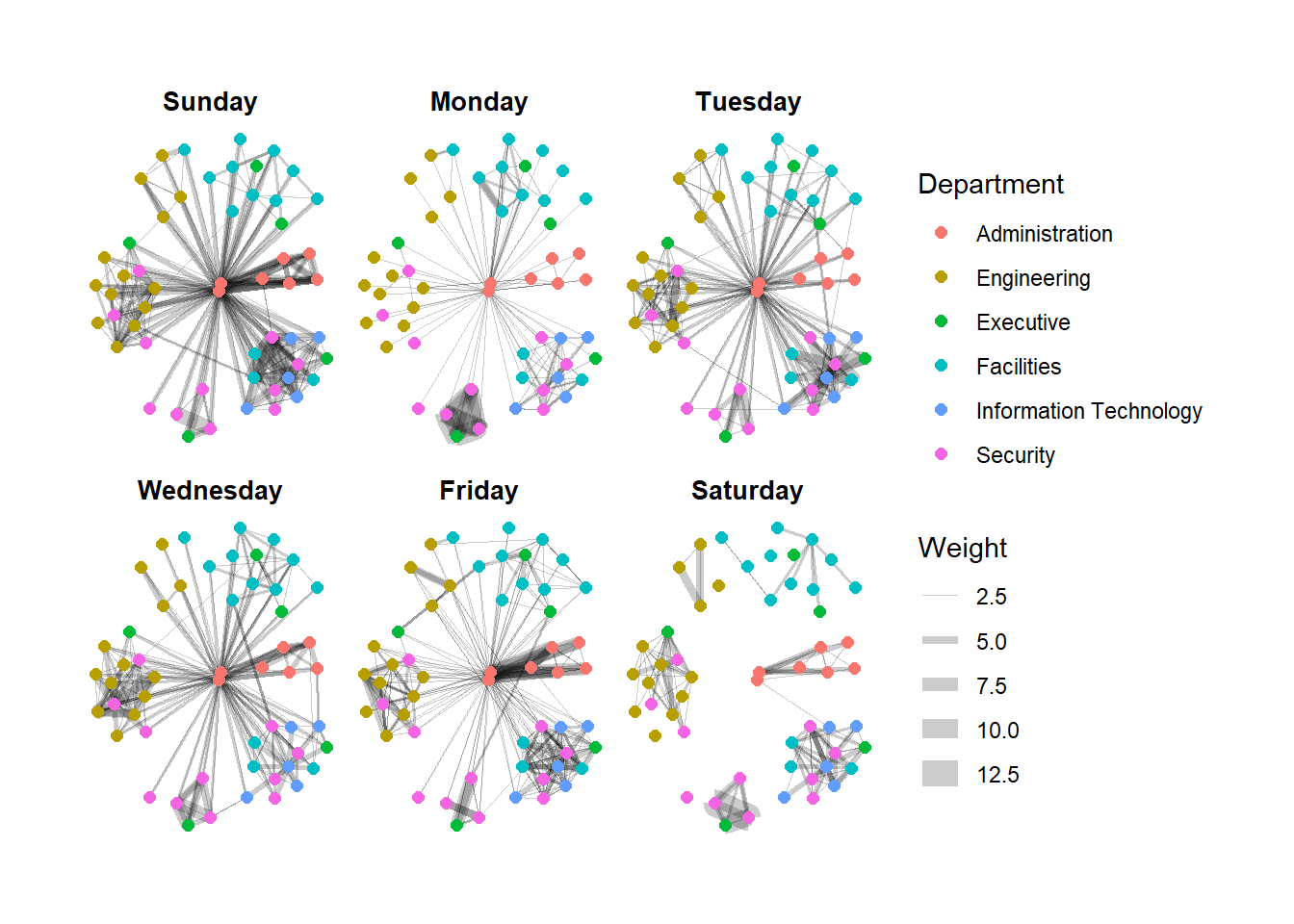
Working with facet_nodes()
set_graph_style()
g <- ggraph(GAStech_graph,
layout = "nicely") +
geom_edge_link(aes(width=Weight),
alpha=0.2) +
scale_edge_width(range = c(0.1, 5)) +
geom_node_point(aes(colour = Department),
size = 2)
g + facet_nodes(~Department)+
th_foreground(foreground = "grey80",
border = TRUE) +
theme(legend.position = 'bottom')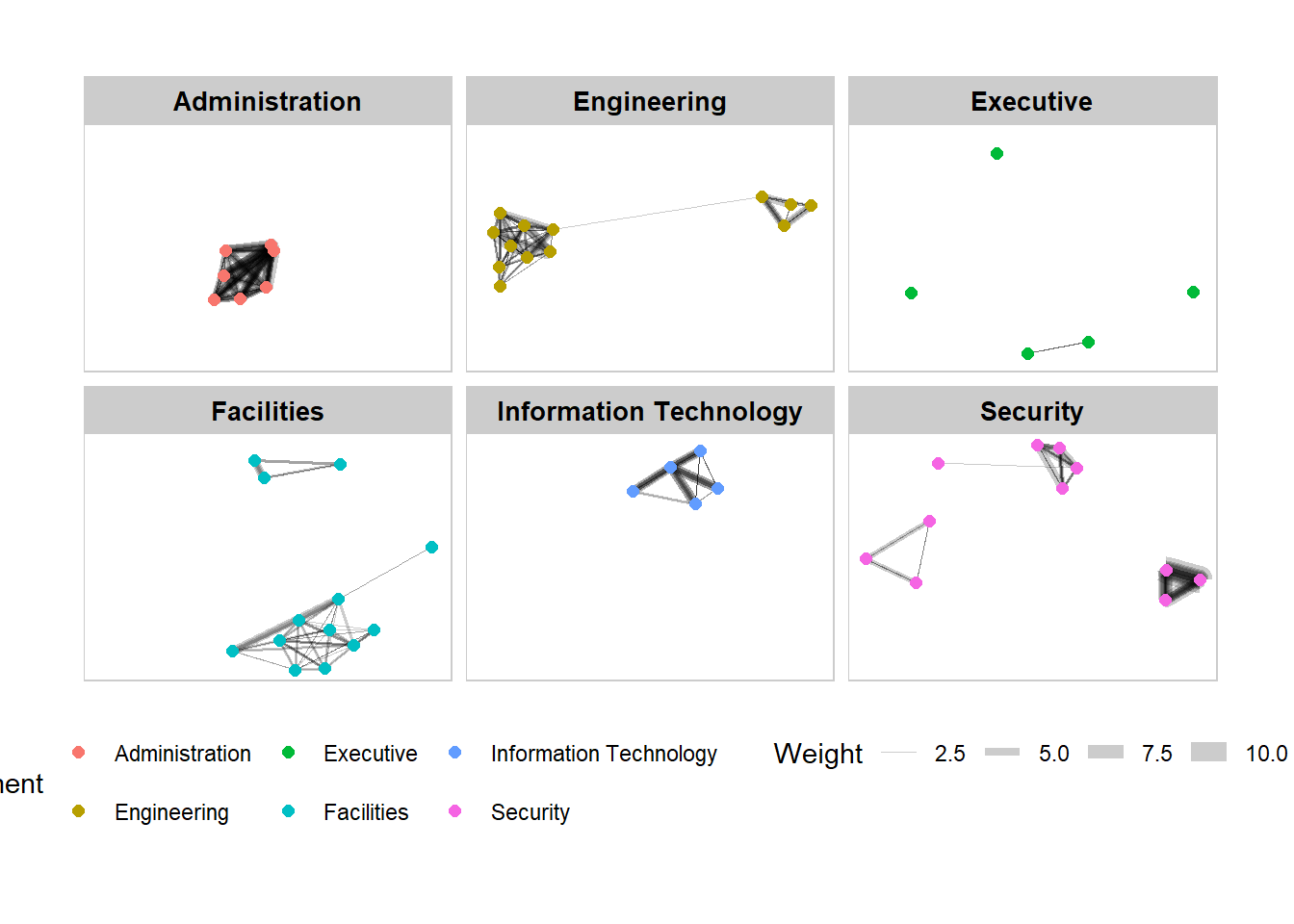
Computing Centrality Indices
g <- GAStech_graph %>%
mutate(betweenness_centrality = centrality_betweenness()) %>%
ggraph(layout = "fr") +
geom_edge_link(aes(width=Weight),
alpha=0.2) +
scale_edge_width(range = c(0.1, 5)) +
geom_node_point(aes(colour = Department,
size=betweenness_centrality))
g + theme_graph()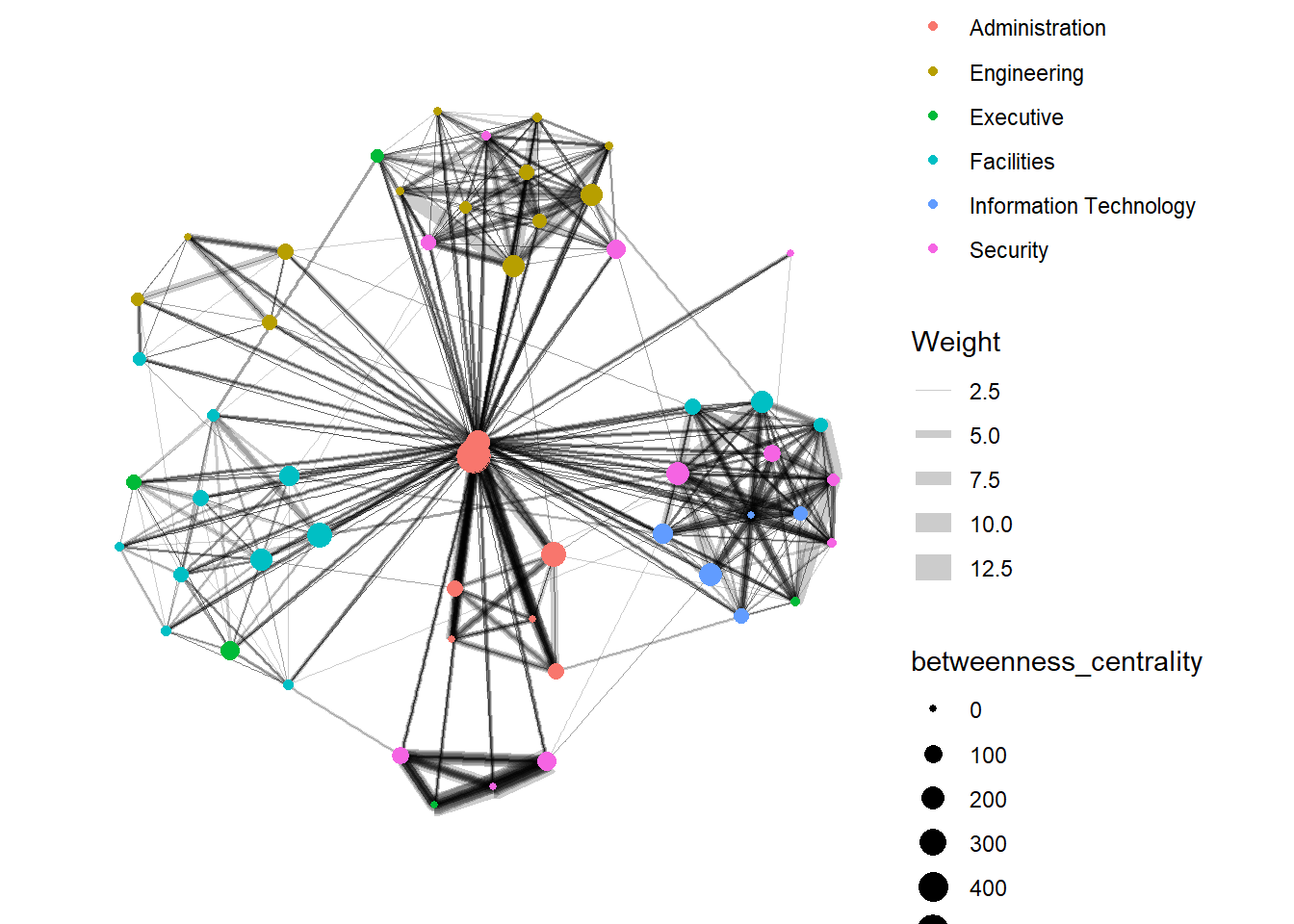
Building Interactive Network Graph with visNetwork
Data Preparation
Before we can plot the interactive network graph, we need to prepare the data model by using the code chunk below.
GAStech_edges_aggregated <- GAStech_edges %>%
left_join(GAStech_nodes, by = c("sourceLabel" = "label")) %>%
rename(from = id) %>%
left_join(GAStech_nodes, by = c("targetLabel" = "label")) %>%
rename(to = id) %>%
filter(MainSubject == "Work related") %>%
group_by(from, to) %>%
summarise(weight = n()) %>%
filter(from!=to) %>%
filter(weight > 1) %>%
ungroup()Plotting the Interactive Network Graph with visual attributes - Nodes
The code chunk below rename Department field to group.
GAStech_nodes <- GAStech_nodes %>%
rename(group = Department) In the code chunk below, Fruchterman and Reingold layout is used.
visNetwork(GAStech_nodes,
GAStech_edges_aggregated) %>%
visIgraphLayout(layout = "layout_with_fr") %>%
visLegend() %>%
visLayout(randomSeed = 123)